Documentation
User Guide
Data upload and metadata capture
Note: Given that DataFlow is currently supported only by the Globus data adapter, the following pointers would focus on this data adapter.
Web interface
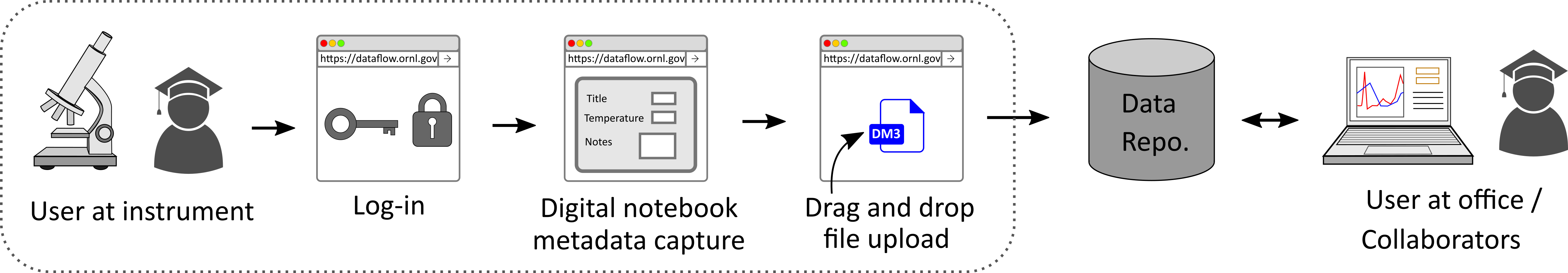
The graphic above provides a high-level view of the key steps in using DataFlow via the web interface.
Regardless of the kind of deployment (central or facility-local), users start to use DataFlow by logging into DataFlow using their XCAMS / UCAMS credentials.
Once authenticated, you will most likely be prompted for UCAMS / XCAMS credentials again, this time to activate the Globus Endpoints between which data are transferred.
Please see instructions below for changing the destination for the data transfers. The default is the user's Home directory in CADES' Open Research file system.
At this point, users are free to click on the New Dataset button to start the process of capturing metadata and uploading raw data associated with a single experiment. Once the New Dataset button is clicked, users will be presented with a panel where they can enter metadata regarding their current experiment, starting with a Title.
- After this, they may select the instrument they are working on (if relevant). If a valid Instrument were selected, a handful of metadata fields would appear per the specifications of the instrument super-user's request.
- Required fields would be marked with an asterisk, highlighting that users must fill in valid values for those fields.
- Users are welcome to add their own fields and enter any metadata they are interested in recording.
Upon clicking the Next button on this panel, the user would now be asked to specify the data files they wish to upload from the computer they are working on.
Users can either select files or folders to upload by clicking on the Upload File or Upload Folder buttons.
Alternatively, users are welcome to drag and drop files or folders into the upload window.
Note:
- Chosen or dragged-and-dropped files / folders will be uploaded immediately upon selection. There is no need to click any other button to upload
- Users should avoid naming their data files as metadata.json since this name is reserved for writing any captured metadata.
As and when files or folders are selected, users will see a blue progress bar indicating the progress in uploading these dataset to the destination.
Once the files are uploaded, users are free to close the file-upload window.
Users can repeat steps 4-5 for subsequent experiments.
Programming interfaces
Before beginning, make sure to follow the steps in the Getting Started page to generate the API key, generate encrypted versions of your password(s), and setup the programming environment of your choice.
The python interface to the REST API mimics the REST API almost exactly. Therefore, users are welcome to use the quick walkthrough as a guide to using either the REST or python interface to DataFlow.
The basic steps that are recommended to upload a dataset with metadata are:
- Make sure to communicate with the appropriate DataFlow server (facility-local / central deployment) in the API URL
- Create an instance of API class if using the python interface.
- Verify and/or change the default settings (destination Globus endpoint for now) using the user-settings REST API calls or settings_get() and settings_set() functions. Please see additional instructions below for changing the default destination.
- Verify that the Globus endpoints are active using the transports/globus/activation REST API call or its python equivalent - globus_endpoints_active().
- Activate the source and destination endpoints using the transports/globus/activate REST API call or its python equivalent - globus_endpoints_activate().
- Create a dataset with appropriate title and scientific metadata using the datasets REST API call or dataset_create() python function.
- Upload a data file to this dataset using the dataset-file-upload REST endpoint or the file_upload() python function.
Users are recommended to use combinations of these functions / API calls to accomplish bigger tasks.
Note that DataFlow currently does not have a mechanism to track progress / completion of a file upload. It is safe to assume that if you get a safe response from DataFlow, then the transfer was at least successfully initiated.
Configuring the destination
By default, data are transferred to the users' HOME directory in CADES' Open Research (OR) Network File System. However, DataFlow allows users to change the destination Globus endpoint.
Finding alternate Globus endpoints
Most high performance computing and data facilities have a Globus Endpoint (or more) that allow one to access their file-system(s). You can also set up Globus endpoint(s) on your personal computer(s) using Globus Personal Connect.
Once you have identified the endpoint you'd like to use:
- Visit https://globus.org
- Find the endpoint that points to the file-system you would like to use as your destination using the "Collection" search bar in the "File Manager" panel. Do not select the endpoint just yet.
- Click on the three vertical dots on the right hand side next to the endpoint of interest.
- The string next to "Endpoint UUID" is the unique identifier for this endpoint. Use this to specify the destination Globus Endpoint in DataFlow
Specifying alternate Globus endpoints
Web Interface 1. Once logged into DataFlow, click on your name on the top right of the screen 2. Select the "Settings" option 3. You should be able to see a box underneath the Destination Endpoint ID option. Paste the Globus Endpoint's UUID there.
Programming interfaces
- REST API - Use the user-settings REST API POST call to change the default destination
- Python - Use the settings_set() function to reset the Globus endpoint.
Accessing uploaded data
By default, data are transferred to the users' HOME directory in CADES' Open Research (OR) Network File System. If you changed the default destination endpoint, you can still follow the same steps but you would need to change how you access / address the file system.
Users can acess this filesystem through multiple ways. Here we list two:
- Globus' web application:
- Log into https://globus.org
- Select the "File manager" from the menu on the left-most column
- Search for the CADES#CADES-OR in the "Collection" search box
- Activate this endpoint by authenticating with your UCAMS / XCAMS credential
- You should be able to the contents of your Home directory
- Logging into the CADES SHPC Condos cluster
- Open a terminal / Powershell
- Log into the cluster via ssh <ucams ID>@or-slurm-login.ornl.gov. Swap with ssh with scp or sftp for transferring data
DataFlow uploads data according to the following nomenclature:
/~/dataflow/<Scientific_Instrument_Name>/<YEAR>-<MONTH>-<DATE>/<HOUR>-<MIN>-<SEC>-<Dataset_Title>/
As an example
/~/dataflow/CNMS-SEM-Hitachi/2022-04-28/08-30-12-Titanium_from_GaTech/
Note:
- If no instrument was selected when uploading data, the second directory would be named as unknown-instrument.
- This final directory would contain both the metadata and the raw data files selected by the user.
- The metadata for each dataset is written into a file called metadata.json.
Note
Given that data on the User's HOME on CADES are expected to be stored indefinitely, the allocated capacity per user is expected to be finite. Therefore, we encourage facilities or users interested in using large (> 50 GB) volumes of data to get in touch with CADES for a more scalable storage solution
Data access, transfer, and sharing
- Users are encouraged to read CADES' documentation on moving data.
- We recommend that users read CADES' documentation on Globus
to easily copy data to their personal computers or
other locations.
- CADES' HOME directory can readily be accessed using the CADES-OR endpoint in Globus
- CADES also provides documentation on sharing data with other users using Globus