Documentation
Context
Motivation
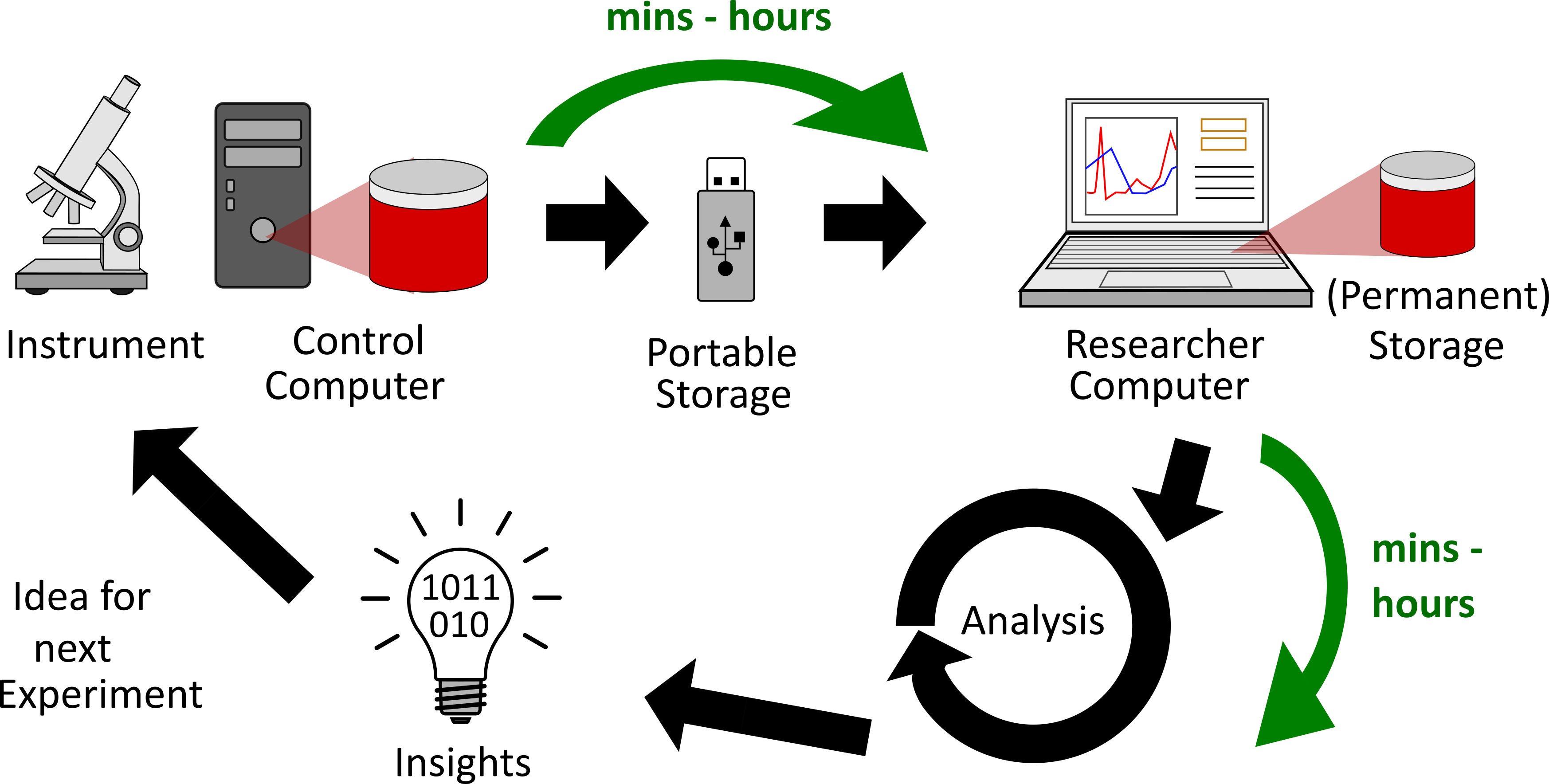
The figure above illustrates the typical iterative process by which experiments are conducted in the observational sciences.
- Creation:
- The researcher performs an experiment using a scientific instrument which in-turn results in data files.
- The researcher also notes down metadata such as the phenomenon or sample being investigated, intent of the experiment, experiment parameters, etc. into a physical notebook.
- Transfer: The data files are copied over from the instrument to the researcher's laptop or workstation via a portable storage drive (e.g. USB thumb drive)
- Analysis:
- Depending on the needs, data are analyzed on the researcher's computer or on a larger machine to gain insights about the current experiment. These insights are used to plan the next experiment (same or different parameters and intents)
- Notes taken down in the physical lab notebook may or may not have been used for the analysis.
- Archive:
- Data remain on the computer(s) where the data were analyzed
- The physical lab notebook is archived in a bookshelf once all pages are filled in.
Gaps and Challenges
- Lost metadata: The vital and rich information in the lab notebook are rarely digitized, married to the raw data to provide a comprehensive view of the experiment, potentially used during the analysis or provided alongside data files when sharing or publishing data.
- Inconsistent metadata: Researchers record metadata in styles that may not match with others within the same lab or even compared to themselves from a day ago. This would make it collected information more challenging to reuse, share, and interoperate.
- Storage capacity:
- Unless data are routinely deleted off the instrument computer, the storage drive in the instrument computer would fill up and preclude future experiments from being conducted using the instrument
- Similarly, the ability to collect, store, and archive data for individual researchers seems to be limited by the capacity of the computers (or external) storage device.
- Catastrophic failure: Unless data on the instrument / personal computer are routinely backed up, researchers run the risk of losing valuable data when the storage drive(s) on the computer(s) fail.
- Slow transfer: It often takes several minutes to hours to copy data from the instrument computer via the standard USB interface. During this time, the instrument may be unusable.
- Manual analysis: Currently, the user has to drive the analysis software manually to analyze any data being generated by the instrument.
- Non-scalable analysis: Given the challenges in moving the data, researchers today are limited to their personal computers or workstation for analyzing data between experiments. Other resources like cloud computing or high performance computing are out of reach for most scientists thereby extending the time necessary to gain the insights necessary to inform the plans for the next experiment.
- Off-network instruments: Often, instrumentation software are incompatible with the latest security patches or operating system updates.
Consequently, such instrumentation computers are often kept off the network to avoid security vulnerabilities.
- Often these computers are operated with a single user account with elevated privileges which could accidentally (or maliciously) enable users to delete other users' data. Alternatively, if the data are not well organized, it may be impossible to identify the creator(s) of certain data.
DataFlow is aimed at solving the aforementioned challenges by moving data and metadata where it can be most useful and safe
Overview
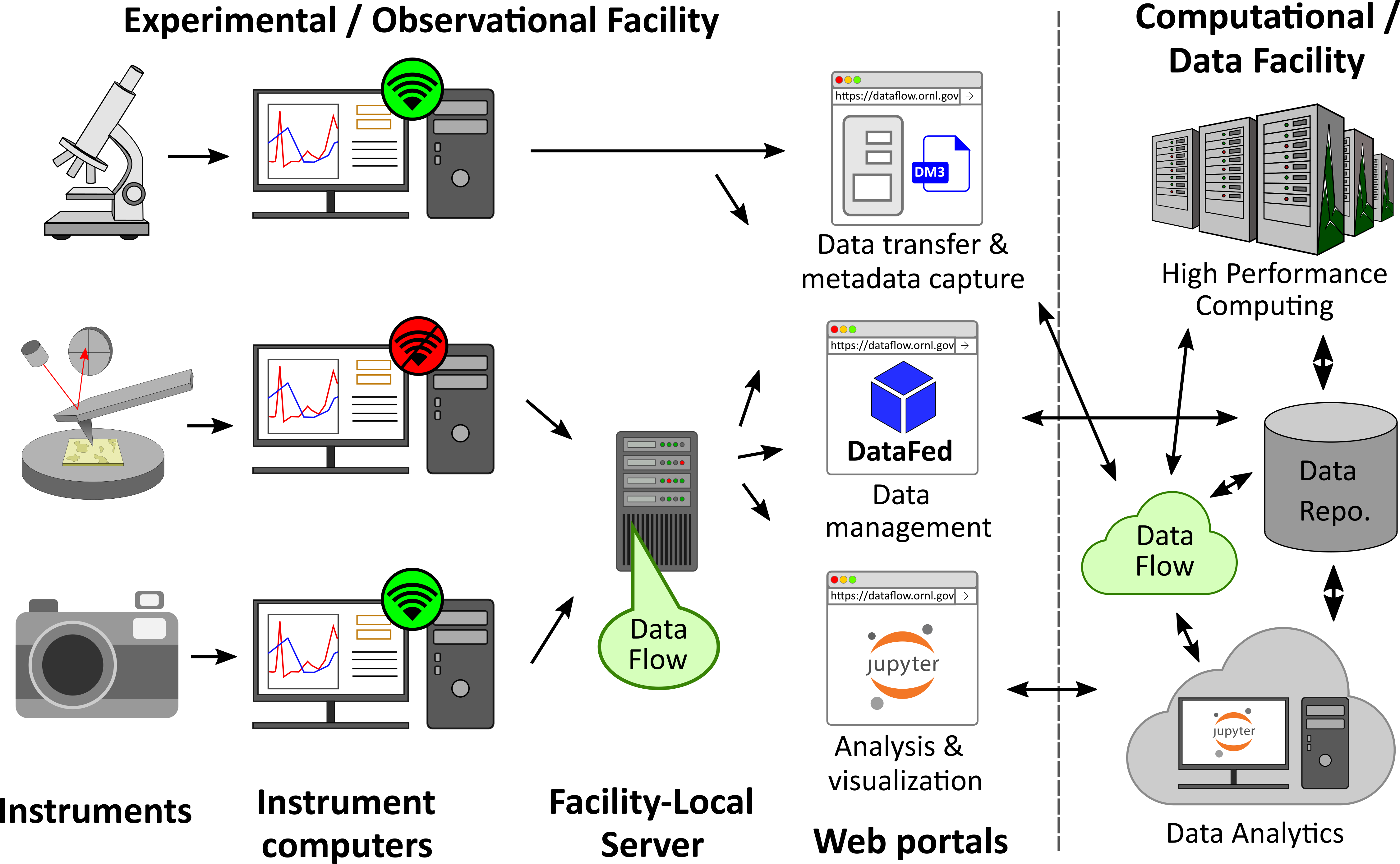
The schematic above provides a high-level overview of the use-cases and capabilities provided by DataFlow.
Primarily, DataFlow is a set of software services and tools that can be accessed either via https://dataflow.ornl.gov (referred to as the "Centralized Deployment") from a networked computer or via a dedicated server installed in close proximity to one or more scientific instruments within a facility, referred to as a "Facility-local Deployment".
Regardless of the deployment mode, users can start using DataFlow by logging in using their ORNL UCAMS / XCAMS credentials. Once authenticated, users can use web forms to enter metadata regarding their experiment as pairs such as "Temperature", and "300". Staff members who manage instruments can develop instrument-specific metadata templates wherein certain metadata fields are pre-populated, mandatory, or optional to ensure consistency in metadata collected from the instrument. Next, users can proceed to upload one or more data files or an entire folder. DataFlow will instantaneously start uploading selected data files to the designated destination. The metadata entered into the web-form will be written into a file named "metadata.json" that will reside next to the chosen data files.
Once data are transferred to the designated storage location, researchers can use existing capabilities in the Compute and Data Environment for Science (CADES) to move data to other locations; use a scientific data management system to share, manage, and organize data; use connected cloud and high performance computing resources for large scale data analytics; publish data, etc.
More information about how DataFlow fits in with the next generation data services is available in this article that was part of the 2020 Smokey Mountains Conference.
Facility-local Deployments
Researchers can request to have the DataFlow software deployed on a dedicated physical server in close proximity to one or more scientific instruments.
Here are some key benefits of / use-cases for facility-local deployments:
- Off-network instruments: Such a deployment would enable instruments that are off the network (such as the scanning probe microscope - center row on the left) to upload data and metadata without requiring the instrument computer to be incorporated into the ORNL enterprise network or be modified in any way. In addition, the dedicated server can also serve as a bridge between off-network instruments and select / approved web-services available elsewhere. As an example, users could manage (organize, share, search, discover) data on a scientific data management system such as DataFed (one of the web services in the center of the image). Another example could be a domain-specific or multi-user data analytics software service such as a JupyerHub (shown in the bottom-right). Such web services could be made available to the researcher without requiring them to leave the instrument computer even if it is otherwise off the ORNL enterprise network.
- Higher / consistent performance: Instrument computers that are already networked (such as the camera on the bottom left), could also use the dedicated server if they are connected to instruments that generate large volumes of data rapidly. Bringing the DataFlow (data movement) services as close as possible to the instrument would ensure that data are transmitted with the highest possible speed to/from the instrument given existing traffic on the network.
- Data pre-processing - (Upcoming capability) The compute power on the server could be used for pre-processing before transmitting the data to the storage repository. Examples of pre-processing include data compression, translating data from proprietary file-formats to community standards, extracting metadata from data files, etc.
- Buffer - (Upcoming capability) The storage on the server could be configured as buffer to alleviate mismatches between the data generation rates and the available bandwidth on the network for situations where data are generated in bursts.
Challenges addressed
- Lost metadata:
- The instrument handler can ensure that metadata are always entered before data are uploaded
- Metadata are always transmitted alongside the raw data
- (Upcoming capability) Should researchers choose (recommended), captured data and metadata could be inextricably tied to each other using a scientific data management system such that information are easily discoverable via simple filtering or complex search queries.
- Inconsistent metadata: The instrument handler can require that key metadata are always captured (and validated) to ensure consistency across users and time
- Storage capacity:
- Economies of scale can be leveraged to provide large capacity (10s of TB to PB) storage for facilities instead of smaller, manually-managed, ad-hoc solutions
- Researchers would not need to worry about storing data on their own computers
- Catastrophic failure (Upcoming capability):
- Periodic backups, even to off-site locations, can be automated.
- Data on instruments could be deleted without worrying about loss if data are consistently being uploaded via DataFlow.
- Slow transfer: Fast and reliable transfers (Globus) using DataFlow
- Manual analysis (Upcoming capability): We plan on using automation technologies to launch automated (standardized) analyses of data using DataFlow. Resultant analyses products could also be injected into the data management system along with provenance links between the source dataset(s) and the product dataset(s).
- Non-scalable analysis:
- DataFlow brings data closer to cloud and high-performance computing thereby lowering barriers to using scalable computing
- (Upcoming capability) We plan on leveraging scalable computing for automated analyses or processing.
- Off-network instruments:
- Data services including DataFlow can be brought to off-network instruments via facility-local deployments without requiring that such instrument computers be modified in any way
- All data services would require users to authenticate with their credentials thereby providing secure access and accountability to data.
Approach
Our aim / philosophy is to develop tools and services that are:
- loosely coupled and modular - We aim to provide researchers with combinations of tools / capabilities they ask such that the the researchers are able to maximize flexibility and freedom in their workflow to suit their unique needs.
- largely domain-agnostic while being sufficiently customizable to each domain without significant effort - By ensuring that the overwhelming majority of our capability is reusable across domains, we are able to develop and maintain a single code-base that is far more robust than multiple-domain specific tools.
- Scalable - We recognize that most scientific domains are experiencing explosions in data volumes, velocity, variety, and complexity. We design our services with scalability, performance, and big / complex data in mind.
Preparing for a shift in workflow
The key philosophy of DataFlow in general is to move information to a location or state that is better than the previous state. Using DataFlow will introduce a cultural shift in the lives of its users requiring them to use new / scalable technologies in place of familiar habits and technologies as the workflows transition from local resources like laptops and USB drives to use scalable resources. It is important note that most aspects in the lifecycle of the dataset, even those further down the data movement stage, will change once information is moved away from instruments.
Here are some examples of changes that DataFlow will likely introduce throughout the lifecycle of a scientific dataset:
- Metadata capture - Researchers would need to conform to standards established at their lab to track metadata into DataFlow's web form rather than note them down in their physical lab notebook
- Data transfer (from instruments) - Data need not be transferred using USB sticks anymore.
- Data availability (from repository) - While it has been convenient to be able to copy data off instruments using portable storage drives, users would now need to use standardized tools to copy data from the centralized repository holding all the data.
- Data analysis - Given that DataFlow moves data away from scientist-managed personal computers towards standardized cloud and high-performance computing resources, the (proprietary) tools traditionally used for analyzing and visualizing data may or may not work on such compute resources. DataFlow lowers the barriers for facilities and groups which have embraced open-source software tools for automating and speeding / scaling up data analysis routines by leveraging high performance compute resources which are far more powerful than conventional workstations. Those who rely on proprietary software or software that only runs on Windows machines may want to either explore alternative software that work on linux operating systems (on CADES resources) or explore the possibility of deploying such software on ITSD administered Windows based cloud infrastructure and moving data closer to those resources using DataFlow. Groups who are unable to transition away from software that only runs on local computers may find DataFlow to be limited to a data archival tool.
- Data storage - Assuming scientists conscientiously use DataFlow to move their data away from instruments, data would either never accumulate at instruments (if users delete their data after uploading them), or can be deleted without worrying about loss from instruments. If users use remote resources like the cloud and high-performance computing resources to analyze and visualize their data, the requirement to download and maintain a copy of all data on personal computers is dramatically reduced.
- Data sharing and management - With DataFlow transmitting data away from local computers and devices, users could use standardized capabilities in the HPC-world to share data with internal and external collaborators. In a future iteration of DataFlow, users would have the option to directly upload data to a scientific data management system which would both enhance and simplify common data management tasks such as sharing, organizing, and searching for data.